We strive to push the boundaries of measurement science and technology for proteins to obtain larger volumes of higher-quality data. The key to unlocking scale and precision is miniaturisation and automation. To achieve this objective, we design new experiments using microfluidic technologies.
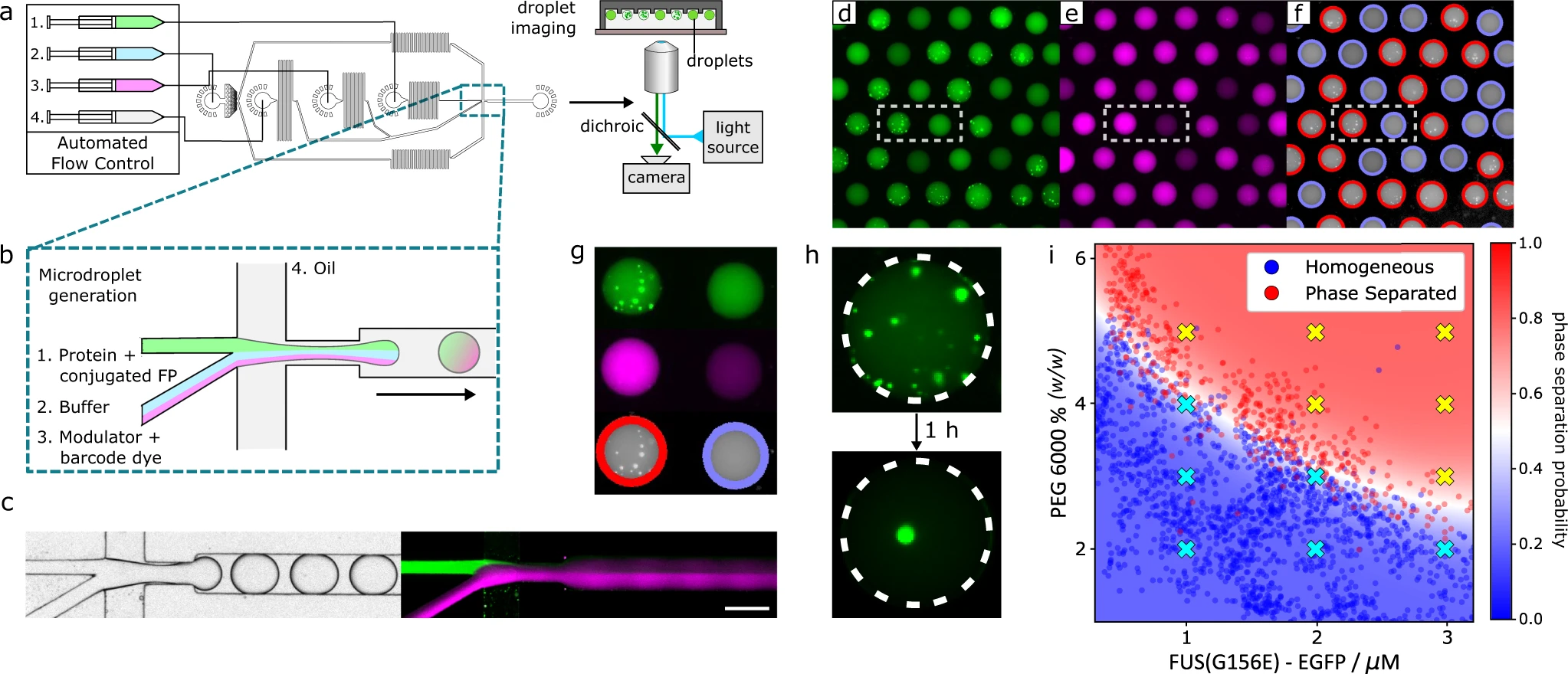
- Arter, W.E., Qi, R., Erkamp, N.A. et al. Biomolecular condensate phase diagrams with a combinatorial microdroplet platform. Nat Commun 13, 7845 (2022). https://doi.org/10.1038/s41467-022-35265-7
- Krainer, G., Jacquat, R.P.B., Schneider, M.M. et al. Single-molecule digital sizing of proteins in solution. Nat Commun 15, 7740 (2024). https://doi.org/10.1038/s41467-024-50825-9
Mechanisms of protein misfolding diseases and therapeutics
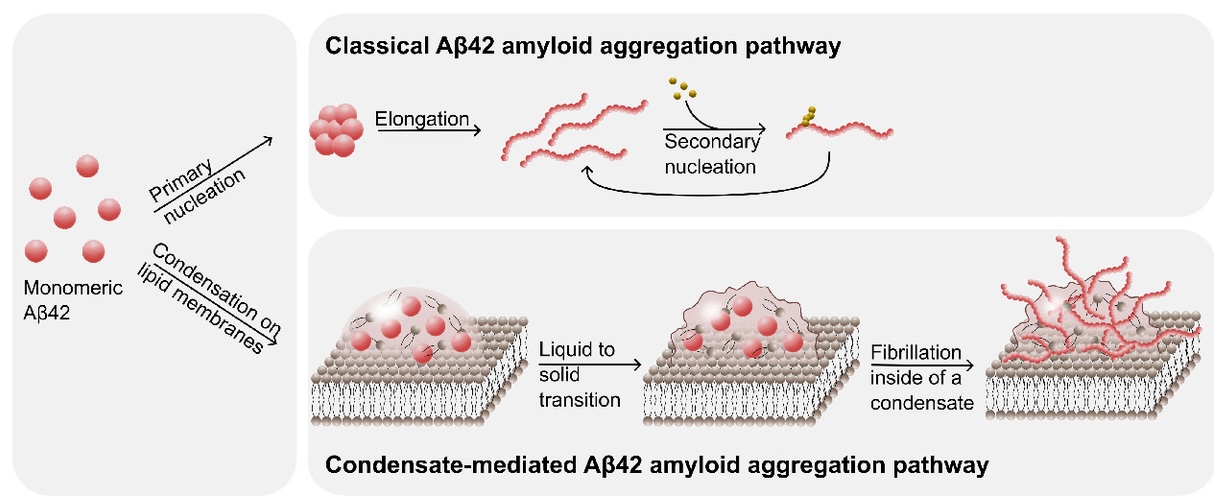
The proper assembly of proteins is crucial for maintaining cellular function. When proteins are misassembled, they lose their intended functionality and can interfere with other cellular processes. This dysfunction is a key factor in the development of various diseases, including Alzheimer’s and Parkinson’s. In recent years, we have made significant progress in uncovering the molecular mechanisms that drive protein aggregation in these systems. Building on this knowledge, we are now focusing on designing more effective therapeutics and investigating the natural defence mechanisms that cells employ to prevent and mitigate harmful protein misassembly.
- Šneiderienė, G., González Díaz, A., Das Adhikari, S., et al. Lipid-induced condensate formation from the Alzheimer’s Aβ peptide triggers amyloid aggregation. Proc. Natl. Acad. Sci. U.S.A. 122, 4 (2025) https://doi.org/10.1073/pnas.2401307122
- Xu, C.K., Meisl, G., Andrzejewska, E.A. et al. α-Synuclein oligomers form by secondary nucleation. Nat Commun 15, 7083 (2024). https://doi.org/10.1038/s41467-024-50692-4
Diagnostics and biomarkers:
The absence of biomarkers to track disease processes and progression has been a major bottleneck, hindering not only the availability of diagnostics for this class of diseases but also complicating the development of therapeutics. These biomarkers are crucial for assessing treatment efficacy and for patient stratification. We are addressing this challenge using new digital microfluidics platforms that we are developing.
Novel materials
Modulating protein assembly gives a route to design new types of biomaterials with applications in medicine and sustainability. Using this strategy, we explore new materials beyond the space used to date by biology, and investigate applications ranging from gene delivery to sustainable coatings.

- Kamada, A., Rodriguez-Garcia, M., Ruggeri, F.S. et al. Controlled self-assembly of plant proteins into high-performance multifunctional nanostructured films. Nat Commun 12, 3529 (2021). https://doi.org/10.1038/s41467-021-23813-6
-
Wiita, E.G., Toprakcioglu, Z., Jayaram, A.K., et al. Formation of Nanofibrillar Self-Healing Hydrogels Using Antimicrobial Peptides. ACS Appl. Mater. Interfaces 16, 46167–46176 (2024). https://doi.org/10.1021/acsami.4c11542
Decoding protein assembly by AI
AI has revolutionised our understanding of protein folding, but comparatively less is known about the rules that govern the interactions and assembly of proteins, especially disordered ones. We are using and developing AI methods to address this challenge, working both with publicly available datasets and data that we generate using our technologies in the laboratory.

- Saar, K.L., Scrutton, R.M., Bloznelyte, K. et al. Protein Condensate Atlas from predictive models of heteromolecular condensate composition. Nat Commun 15, 5418 (2024). https://doi.org/10.1038/s41467-024-48496-7
- Saar, K.L., Morgunov, A.S., Qi, R., et al. Learning the molecular grammar of protein condensates from sequence determinants and embeddings. Proc. Natl. Acad. Sci. U.S.A. 118, e2019053118 (2021). https://doi.org/10.1073/pnas.2019053118